Latest News

Congratulations, Dr. Adrián Contreras!
Congratulations, Dr. Adrián Contreras!

Congratulations, Dr. Benjamin Buchfink!
Benjamin Buchfink defended his thesis today. Congratulations!

Congratulations, Dr. Maximilian Collenberg!
Max successfully defended his thesis today.
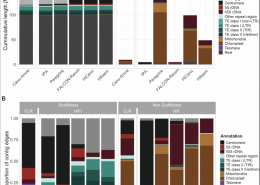
Out in NAR: Pushing the limits of HiFi assemblies (OA)
In-depth comparison of PacBio CLR and HiFi assemblies

Congratulations, Dr. Alejandra Duque!
Alejandra successfully defended her thesis
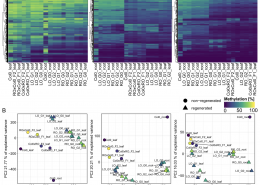
Now in Quant. Plant Biol.: New tool for DNA methlyation analysis
Methylscore to ID differentially methylated regions

Former postdoc Jiawei Wang wins Xplorer Prize
..which comes with a personal cash award of 3M RMB (~430,000€)

Detlef spoke at Kinderuni
Covered in our local newspaper
Tübingen Children's University
Max…
